The probiotic strains Lactobacillus spp. and Bacillus spp. make probiotics in shrimp farming thanks to their beneficial properties such as supporting digestion and inhibiting pathogenic bacteria in shrimp. This study obtained some strains of Lactobacillus spp. and Bacillus spp. that have probiotic properties from the pond of whiteleg shrimp (Litopenaeus vannamei) in Truong Dinh village, Hoa Hiep commune, Da Nang city, as a basis to produce probiotics for shrimp. The bacterial strain B3 with the highest ability to produce amylase and protease enzymes, with a sterile ring diameter of 27 ± 1.4 mm and 25 ± 0.6 mm, respectively, and strain LT2, antagonizing Vibrio parahaemolyticus with a sterile ring diameter of 25 ± 2.12 mm were selected. The 16S rRNA method combined with morphological and biochemical characteristics showed that strain B3 is Bacillus Subtilis and strain LT2 is Lactobacillus Plantarum.
Keywords: Bacillus subtilis, Lactobacillus plantarum, antagonizing, enzyme, probiotics
Problem:
Vietnam is one of the countries with strengths and potential for developing the aquaculture industry in general and shrimp farming in particular. According to the shrimp production plan, in 2022, the shrimp farming area will reach 750,000 ha; black tiger shrimp account for 625,000 ha; white shrimp account for 125,000 ha, and the shrimp production of all kinds is 980,000 tons. Export turnover is over 4 billion USD (up 2.56 % compared to 2021). However, the abuse of antibiotics and disinfectants in aquaculture to prevent and control diseases has caused many serious consequences, affecting the health of consumers (Dorsey & Robertson, 2013). Therefore, probiotics have become one of the new approaches to replace antibiotics.
Probiotics are used to replace antibiotics in seafood, helping to increase the survival and growth of aquatic animals (Reyes-Becerril et al., 2014). Among the probiotic microorganisms, Lactobacillus spp. and Bacillus spp. play an important role in the host's gastrointestinal tract by improving immunity, balancing intestinal microflora, and secreting antibacterial substances such as lactic acid, acetic acid, bacteriocin... which inhibit the growth of malignant bacterium (Ige, 2013). Previous studies have also demonstrated that Bacillus sp. produces antimicrobial peptides, which provide immunostimulatory effects for livestock (Barbosa et al., 2005). Spores of the genus Bacillus have an advantage over vegetative cells because they are stable over long periods, can be produced commercially, and are widely used as a biological agent against pathogens and naturally digested by animals (Hong et al. 2005). Senok et al, 2005 also reported that lactic acid bacteria such as Lactobacilli and Bifidobacteria help lower the pH of the gastrointestinal tract by converting lactose into lactic acid. In this way, the conquest of many malignant bacteriums in the gastrointestinal tract of aquatic animals is prevented. Besides, Verschuere et al., (2000) suggested that Bacillus sp. improve water quality parameters as they are added to the culture system. However, most of the microbiological preparations used in the country today are of the imported origin or unknown ingredients and types. Microbiological products isolated and domestically produced are still limited. The study and selection of indigenous beneficial bacterial strains as the basis for the mass production of microbial products is essential and has practical significance in the current period to improve production efficiency, limit environmental pollution, promote and enhance the sustainability of aquaculture. Therefore, the study was conducted to isolate, select, and investigate the beneficial properties of Lactobacillus spp. and Bacillus spp. from white leg shrimp ponds in Truong Dinh village, Hoa Vang district, Da Nang city, contributing to improving shrimp farming efficiency.
2. Materials and method
2.1 Materials
— 05 intestine samples of healthy whiteleg shrimp ( Litopenaeus vannamei ), 3 slugde samples, and 1 water sample from shrimp pond in Truong Dinh village, Hoa Hiep commune, Da Nang city.
— The test microorganism strain Vibrio parahaemolyticus was provided by microbiological technology laboratory, faculty of Biology — Environment.
2.2 Research methods
2.2.1. Lactobacillus and Bacillus isolation
Lactobacillus isolation: Conduct a dissection to remove the intestines of white leg shrimp. Samples of shrimp intestines are put into test tubes containing 5 ml of sterile physiological saline; grind the sample homogenized with physiological saline to settle by a stick. Dilute the culture solution 10–2, 10- 3, 10–4. Aspirate 50 µl of the sample solution and spread it on MRS agar (+ 1 % CaCO3) and incubate anaerobically at 28ºC for 48 h. Select characteristic colonies with CaCO3halo zone diameter, observe the cells under the microscope and then transplant pure lines in MRS agar and 25 % glycerol (Huong Truong Giang, 2020).
Bacillus isolation : One gram of sludge sample (water sample) mixed in 9 mL of sterile saline (0.85 % NaCl) and heat-treated at 80ºC for 20 min. Then, 100 µL of sample diluent at appropriate concentrations (10–1, 10–2, 10–3) was spread evenly on an LB agar plate and incubated at 28ºC for 24 h. Single colonies were selected and transplanted several times on LB agar plates to obtain pure colonies (Nguyen Lan Dung et al., 1975).
2.2.2.Preliminary bacteria identification by biochemical test
Gram staining : the purity and morphology of bacterial strains were observed by Gram staining according to the Hucker and Conn method(1923).
Catalase response : a sterile culture rod is used to collect a few colonies and spread them on a slide, then apply a drop of H2O2 solution (3 %) to the slide. Bacteria that are positive for catalase will have gas bubbles and vice versa.
O xidase response : spread a few bacteria onto a paper plate impregnated with Tetramethyl — p — Phenylendiamine dihydrochloride solution (1 %) by a sterile culture rod. Bacteria that give a positive reaction will turn the paper black and vice versa.
Indole test: Culture bacteria in 1 % peptone water without sugar and incubate at 37°C. After 24–48 hours, add 10 drops of Kovac's reagent and gently shake the test tube. Observe after 5 minutes that if a red ring appears, the reaction is positive. If yellow appears, it is negative.
M obility test:
Principle: detect the moving ability of bacteria in the culture medium by flagella and flagellum.
Procedure: deeply inoculate biomass into soft agar (0.5 % agar). Incubate 37℃ from 18–24 hours (Nguyen Lan Dung et al., 1975).
Sugar ferment ability : inoculate the bacterial strain into a culture medium containing phenol red carbohydrate broth (trypticase: 1 %; NaCl: 0.5 %; red phenol: 0.018g/l; carbohydrates: 0.1 % (glucose), sucrose, lactose...), cultured at 37ºC/24–48h, microorganisms using sugar sources in the medium will reduce pH, change the color of red phenol indicator. Result: (+): medium turns yellow; (-): the medium is red (Nguyen Lan Dung et al., 1975),
2.2.3. Evaluation of extracellular enzyme activity
Bacterial strains of Bacillus spp. and Lactobacillus spp. were grown on LB and MRS medium. After 24 hours, centrifuge at 6000 rpm for 10 minutes to remove bacterial cells and collect the supernatant to determine protease and amylase enzyme activities.
The proteolytic ability of bacteria was evaluated by its ability to form halo zone diameter in Casein medium (10g/l casein; 0.5g/l KCl; 1.5g/l K2HPO4; 0.5g/l MgSO4. 7H2O; 0.01g/l FeSO4.7H2O; 20g/l agar)
The amylase activity was determined by the ability to form lytic rings on soluble starch medium including 1g/l K2HPO4; 0.5g/l MgSO4.7H2O; 0.5g/l KCl; 0.01g/l FeSO4.7H2O; 2g/l NaNO3; 10g/l Soluble starch; 20g/l agar
2.2.4. Evaluation of the ability to antagonize Vibrio parahaemolyticus
In order to select strains of Lactobacillus spp. and Bacillus spp. resistant to Vibrio parahaemolyticus . The perforation method (Schlinger & Lücke, 1989) determined the antibacterial activity. NA agar was spread with V. parahaemolyticus at a 105 CFU/ml density and then created agar holes. Aspirate 100 µl of the culture aliquot of each bacterial strain into the agar holes and incubate at 37°C. After 24 hours, measure the antibacterial ring diameter (∆D). ∆D = D — d (mm) with D: diameter of antibacterial ring (mm); d: diameter of agar hole (mm)
2.2.5. Identification method of Lactic acid bacteria and Bacillus bacteria .
Identification by molecular biology method at Nam Khoa company, address: 793/58 Tran Xuan Soan, Tan Hung Ward, District 7, Ho Chi Minh City. Amplification and sequencing of the 16S rRNA region isolated from the bacterial genome. Compare and identify strains using NCBI's online BLAST program.
3 . Research result
3.1. Lactobacillus and Bacillus isolation result
From soil, water, and shrimp gut samples isolated on MRS and LB medium, 5 different bacterial strains were obtained. Based on the microscopic observation of cell shape, biochemical and physiological characteristics, and using the classification key of Bergey et al. (1957), there are two strains of Lactobacillus spp symbols (symbol LT1 and LT2) and 3 strains of belonging to the genus Bacillus (symbol B1→B3). The results are shown in Figure 1, Table 1, and Table 2.
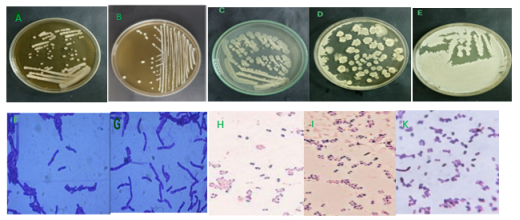
Fig. 1: Colony and cellular morphology of bacterial strains isolated from water, soil, and shrimp gut samples. A, B colonies of strains (LT1, LT2); C- E colonies of strains B1, B2, B3; F-G cells of LT1, LT2, and H-K are cells and spores of B1, B2, B3
Table 1
Morphological characteristics of isolated bacterial strains
|
Symbol |
Isolated source |
Colony characteristics |
Cellular characteristics |
|
|
Shape |
Gram |
|||
|
LT1 |
Shrimp intestine |
White, round, convex colonies with sizes from 1 to 2 mm, capable of dissolving CaCO3 |
Rod cells, arranged in chains, without spores |
+ |
|
LT2 |
Shrimp intestine |
Milky white, round colonies 3–5 mm in size can dissolve CaCO3. |
Rod cells, arranged in chains, without spores |
+ |
|
B1 |
Slugde |
White colonies with saw-line edges |
Rod-shaped, with spores |
+ |
|
B2 |
Water |
White, round colonies with a smooth surface |
Rod-shaped, with spores |
+ |
|
B3 |
Water |
Opaque, convex, wet colonies |
Rod-shaped, with spores |
+ |
|
Note: (+) Positive gram |
||||
Table 2
Biochemical characteristics of 5 isolates of bacteria
|
Quotas |
LT1 |
LT2 |
B1 |
B2 |
B3 |
|
|
Oxidase |
- |
- |
+ |
+ |
+ |
|
|
Catalaza |
- |
- |
+ |
+ |
+ |
|
|
Idol |
- |
- |
- |
- |
- |
|
|
Movability |
- |
- |
+ |
+ |
+ |
|
|
Fermentation ability |
Glucose |
+ |
+ |
+ |
+ |
+ |
|
Maltose |
+ |
+ |
+ |
+ |
+ |
|
|
Lactose |
+ |
+ |
+ |
- |
+ |
|
|
Capability to resolve CaCO3 |
+ |
+ |
- |
- |
- |
|
Note: (+) positive; (-) negative
3.2Investigate beneficial properties of bacterial isolates
Most of the isolated Bacillus strains are capable of producing amylase and protease enzymes. Among them, strain B3 has the most substantial ability to degrade starch and protein with a ring radius of 27 ± 1.4 mm; 25 ± 0.6 mm, respectively (Table 3 and Figure 2). According to another study by Lee et al. (2012), when investigating the probiotic potential of 4 strains of Bacillus spp. isolated from different sources, only the Bacillus sp. SM2 fully expressed amylase, cellulase, and protease enzyme activities, while in the remaining 3 strains, only Bacillus sp. T4 and Bacillus sp. JSP1 exhibited protease activity, and none of these three lines exhibited cellulase activity.
Table 3
Resistance to Vibrio parahaemolyticus and ability to produce extracellular enzymes of isolated bacteria strains
|
Symbol |
Diameter of the inhibition zone (mm) |
Halo zone diameter (mm) |
|
|
Amylase |
Protease |
||
|
LT1 |
20,67 ± 2,08 mm |
- |
12 ± 0,4 mm |
|
LT2 |
25 ± 2,12 mm |
- |
- |
|
B1 |
15 ± 2,4 mm |
21 ± 0,8 mm |
13 ± 1,3 mm |
|
B2 |
12 ± 1,5 mm |
16 ± 1,2 mm |
10 ± 2,4 mm |
|
B3 |
17 ± 2,2 mm |
27 ± 1,4 mm |
25 ± 0,6 mm |
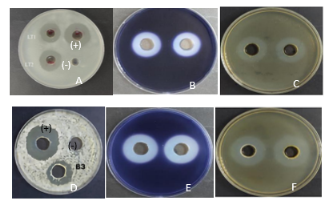
Figure 2. Antimicrobial activity of strains LT1, LT2, B3 against Vibrio parahaemolyticus and amylolytic rings, casein of strains LT2, B3. A-C: Anti-Vibrio parahaemolyticus, amyloid, and casein-degrading activities of strain LT2. D- F: Activity against Vibrio parahaemolyticus , starch, and casein breakdown of strain B3
When testing the ability to antagonize with Vibrio parahaemolyticus , all five strains of bacteria were capable of antagonism. However, strain LT2 gave the highest antagonistic effect at 25 ± 2.12 mm. If we compare our results with Nguyen Thi Truc Linh's study (2016), the results of this study are higher. According to the author, all strains of lactic acid bacteria isolated in Soc Trang have the antibacterial ability, but the antibacterial ring is not significant (11–15 mm).
From the above results, strains B3 have a high ability to secrete extracellular enzymes, while LT2 strains are resistant to Vibrio parahaemolyticus . We carried out these two strains' identification by molecular biology techniques for further studies.
3.3. Results of selected bacterial strains identification
Sequencing results of 16S rRNA genes of strains LT2 and B3 are presented in Figure 3.
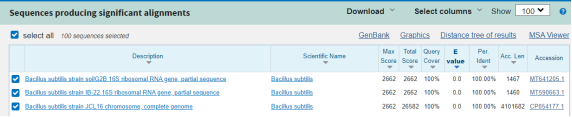

Figure 3. Results of searching for homologous sequences of bacterial strain B3 and LT2
Using the Blast tool on the NCBI website to search for similarity sequences, bacterial strain B3 had a 16S rRNA gene region sequence that was 100 % similar to the species, concluding that strain B3 was Bacillus subtilis . Strain LT2 has a 16S rRNA gene region sequence 98 % similar to that of Lactobacillus Plantarum .
Thus, through the identification step at the species level, bacterial strains B3 are Bacillus subtilis , and LT2 are Lactobacillus Plantarum .
- Conclusion:
The study isolated 2 strains of Lactobacillus spp. (LT1, LT2) and 03 strains of Bacillus spp. (B1, B2, B3) from intestine samples of white leg shrimp ( Litopenaeus vannamei ), water and sludge samples from shrimp ponds in Truong Dinh village, Hoa Hiep city, Da Nang city. In which strain B3 has the highest ability to produce amylase and protease enzymes, with a ring diameter of 27 ± 1.4 mm and 25 ± 0.6 mm, respectively, strain LT2 can antagonize Vibrio parahaemolyticus with a sterile ring diameter of 25 ± 2.12 mm. The 16S rRNA method combined with morphological and biochemical characteristics showed that strain B3 is Bacillus subtilis and strain LT2 is Lactobacillus plantarum .
References:
- B. A. Ige, “Probiotics use in intensive fish farming”, Afr. J. Microbiol. Res. 7, pp. 2701–2711, 2013.
- Barbosa et al, (2005), Screening for Bacillus Isolates in the Broiler Gastrointestinal Tract, Applied and Environmental Microbiology 71(2): 968–78.
- Claus D et al, (1986), Genus Bacillus Cohn 1872. In: Sneath PHA, Mair NS, Sharpe ME, Holt JG (eds) Bergey’s, Manual of Systematic Bacteriology, vol 2. Williams & Wilkins, Baltimore, pp 1104–113.
- D. Dorsey, W. Robertson, “Recent advances in fish diseases treatment: probiotics as alternative therapy to antibiotics in aquaculture”, Eur. J. Ocean Mar 11, pp. 20–28, 2013.
- Hong, H.A., L. H. Duc, and S. M. Cutting., 2005. The use of bacterial spore forms as probiotics. FEMS Microbiol. Rev. 29: 750–757
- Hucker et al (1923), Methods of Gram Staining, New York State Agricultural Experiment Station Technical Bulletin.
- Huynh Truong Giang, Nguyen Hoang Nhat Uyen, Vu Hung Hai, Pham Phi Tuyet Ngan and Vu Ngoc Ut. Evaluation of lactobacillus activity from the intestine of white shrimp with probiotic potential added to shrimp feed. Scientific Journal of Can Tho University, Vol. 56, Special issue: Fisheries (2020) (1): 102–111
- Lee, J., Park, I., Choi, Y. and Cho, J., 2012. Bacillus strains as feed additives: In vitro evaluation of its potential probiotic properties. Revista Colombiana de Ciencias Pecuarias, 25: 577–585
- M. Reyes-Becerril, F. Ascencio, V. Gracia-Lopez, M. E. Macias, M. C. Roa, M. A. Esteban, “Single or combined effects of Lactobacillus sakei and inulin on growth, nonspecific immunity and IgM expression in leopard grouper (Mycteroperca)
- Nguyen Lan Dung, Pham Van Ty and Duong Duc Tien, 1975. Microbiology, University and Professional High School Publishing House, volume 1, 219 pages.
- Nguyen Thi Truc Linh, Nguyen Trong Nghia, Dang Thi Hoang Oanh and Truong Quoc Phu, 2017. Effect of lactic acid bacteria added to feed on resistance to acute hepatopancreatic necrosis disease in white leg shrimp ( Litopenaeus vannamei ). Science Journal of Can Tho University. 52B: 122–130.
- Senok et al, (2005), Probiotics: Facts and Myths, Clinical Microbiology and Infection 11(12): 958–66.
- U. Schillinger, F. K. Lücke, “Antibacterial activity of Lactobacillus sakei isolated from meat”, Applied and Environmental Microbiology, 55, pp. 1901–1906, 1989
- Verschuere et al (2000), Probiotic Bacteria as Biological Control Agents in Aquaculture, Microbiology and Molecular Biology Reviews 64(4): 655–71.







